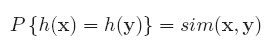First, the objective of learning is formalized as minimizing errors in classification of document pairs, rather than minimizing errors in ranking of documents.Second, the training process is computationally costly, as the number of document pairs is very large. Third, the assumption of that the document pairs are generated i.i.d. is also too strong. Fourth, the number of generated document pairs varies largely from query to query, which will result in training a model biased toward queries with more document pairs.
Thus, in this paper, they propose the list-wise approach.
The followings are the problem setting.
In training, a set of queryies Q=(q(1),q(2), ... ,q(m)) is given.
Each query q(m) is associated with a list of documents d(i) = (d(i)1, d(i)2, ...,d(i)n(i)).
Each list of documents d(i) is associated with a list of judgments (scores) y(i)= (y(i)1, y(i)2, ...,y(i)n(i)). The judgment y(i)j represents the relevance degree of d(i)j to q(i).
A feature vector x(i)j=ψ(q(i),d(i)j) is created from each query-document pair.
Note that there is a q(i) in ψ function. If it doesn't have this item, then the ranking list will be always the same no matter what query is.
We want to find a ranking function f, which will output the score by taking one feature vector.
For the list of feature vectors x(i), we can obtain a list of scores z(i) = (f(x(i)1), ... ,f(x(i)n(i))).
The objective of learning is formalized as minimization of the total losses with respect to the training data.
Where L is the loss function.
In order to measure the loss function of two different ranking, they first convert the ranking into probability distribution, and measure the loss function by using Cross Entropy.
They use the following definition to convert the ranking into probability distribution.
However, if we want to derive this probability distribution precisely, we need lots of computation overhead.
Thus, instead of calculating this probability, they calculate top one probability.
Then, for top one probability, the value can be derived more easily.
After using this representation, then we can use Cross Entropy as metric.
And phi-function is defined as exponential function.
Then, they use gradient descent method to solve this optimization problem.
The ranking function fw is based on the Neural Network model w.
Given a feature vector x(i)j, fw(x(i)j) assigns a score to it.
Thus, for a query q(i), the score list z(i)(fw) = (fw(x(i)1), fw(x(i)2), ..., fw(x(i)n(i))).
For simplicity, they use linear Neural Network model in the experiment, fw(x(i)j) = w × x(i)j.
That is, they tried to learn a linear combination that minimize the loss function measure by Cross Entropy.
And they show their list-wise method really performs better than other methods